Current Research (PhD, 2022–Present)
RESOLFT time lapse imaging empowered by deep learning
Guillaume Minet, Anirban Ray, Francesca Pennacchietti, Giovanna Coceano, Florian Jug, and Ilaria Testa
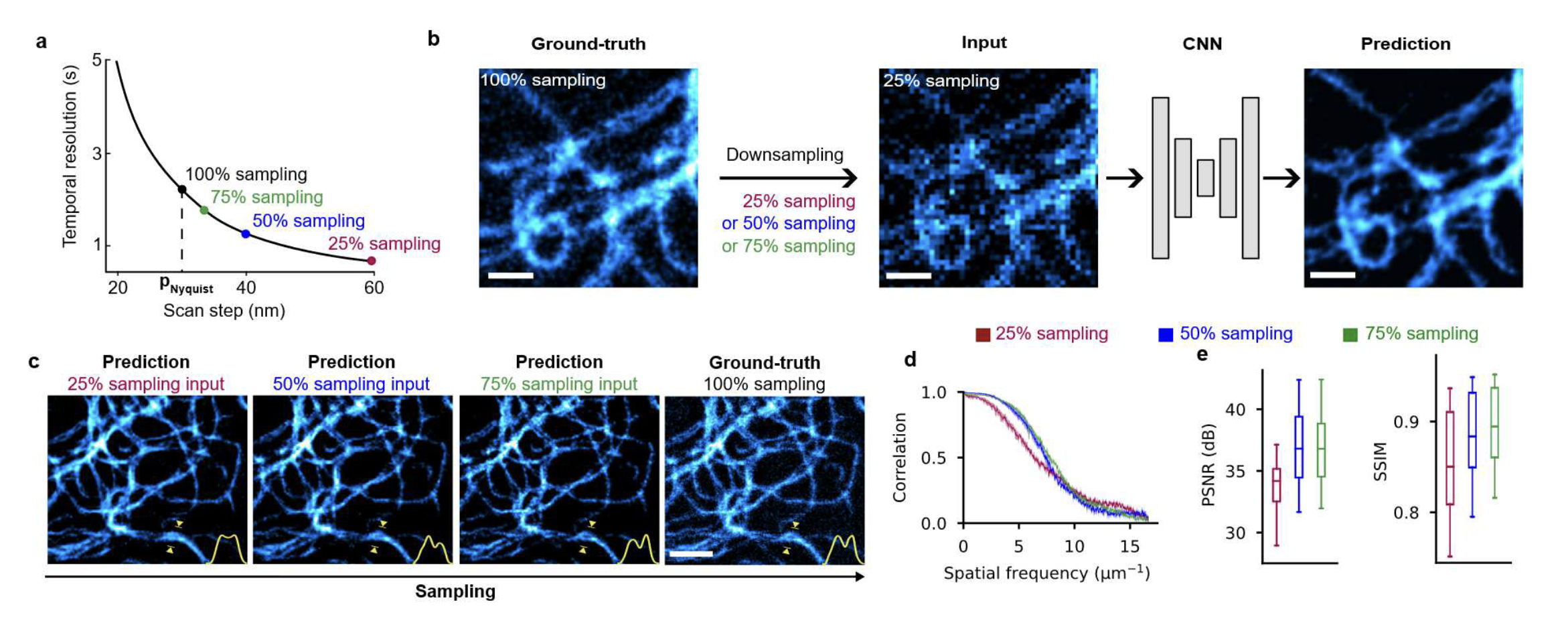
Deep learning extended RESOLFT nanoscopy by restoring low-SNR and sub-sampled acquisitions, enabling 5× longer imaging with 10× lower dose of light per frame, or a 4× increase in imaging speed for faster live-cell imaging while preserving ~60 nm resolution. This method enables reduced photobleaching and accelerated volumetric recording, revealing previously inaccessible sub-organelle dynamics in living cells.
Preprint (under review) | AI Generated infographic 
Past Research (Hitachi Ltd., 2018–2021)
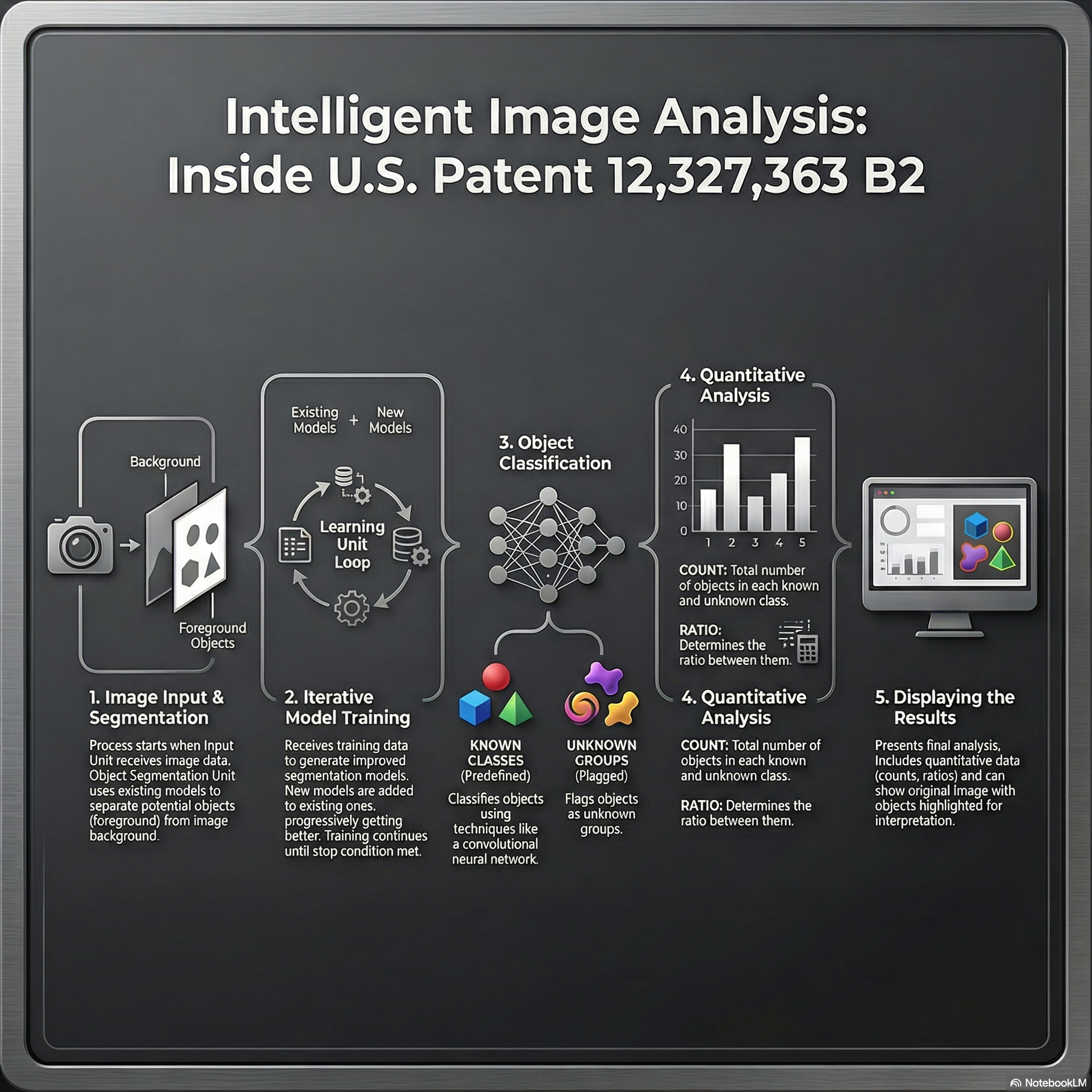
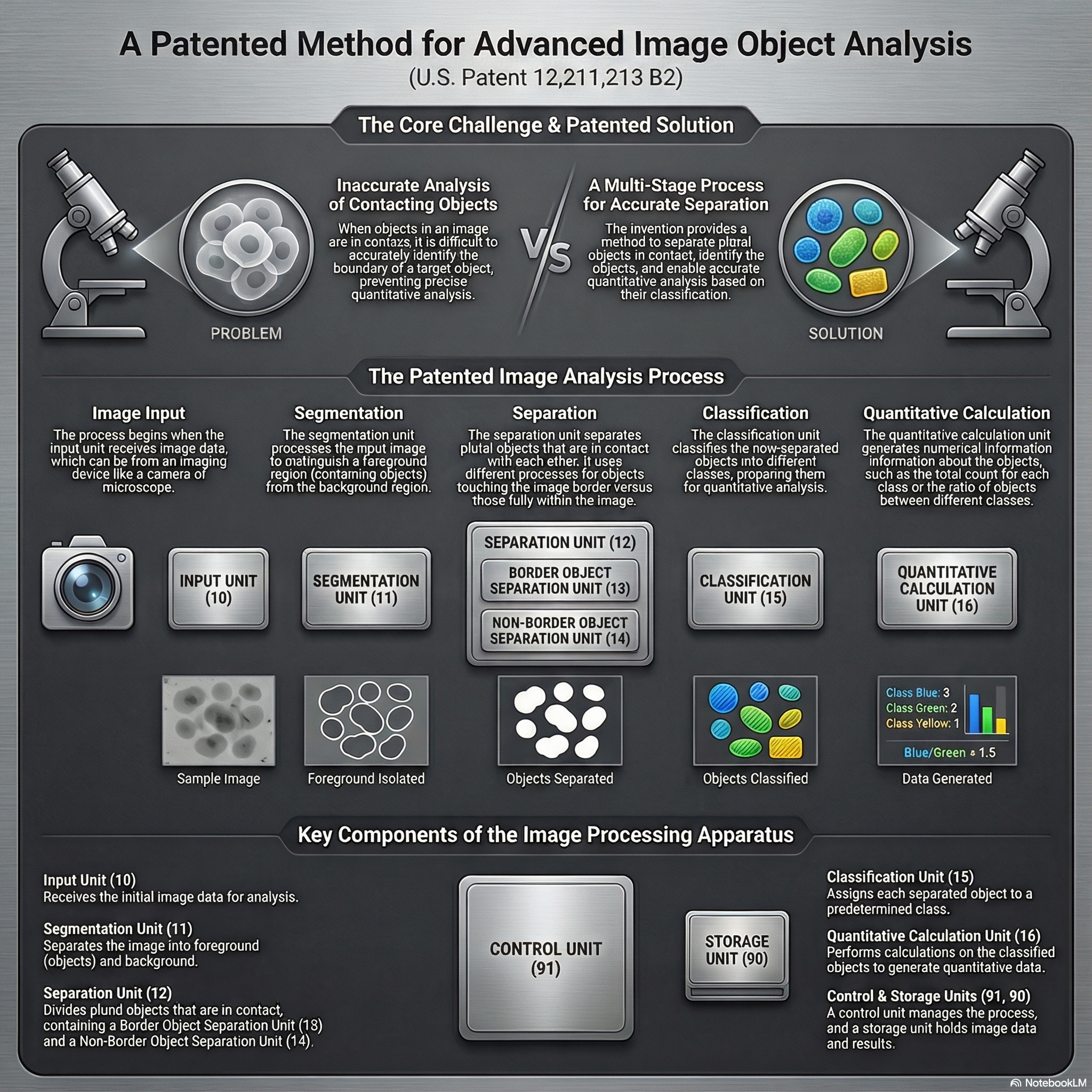
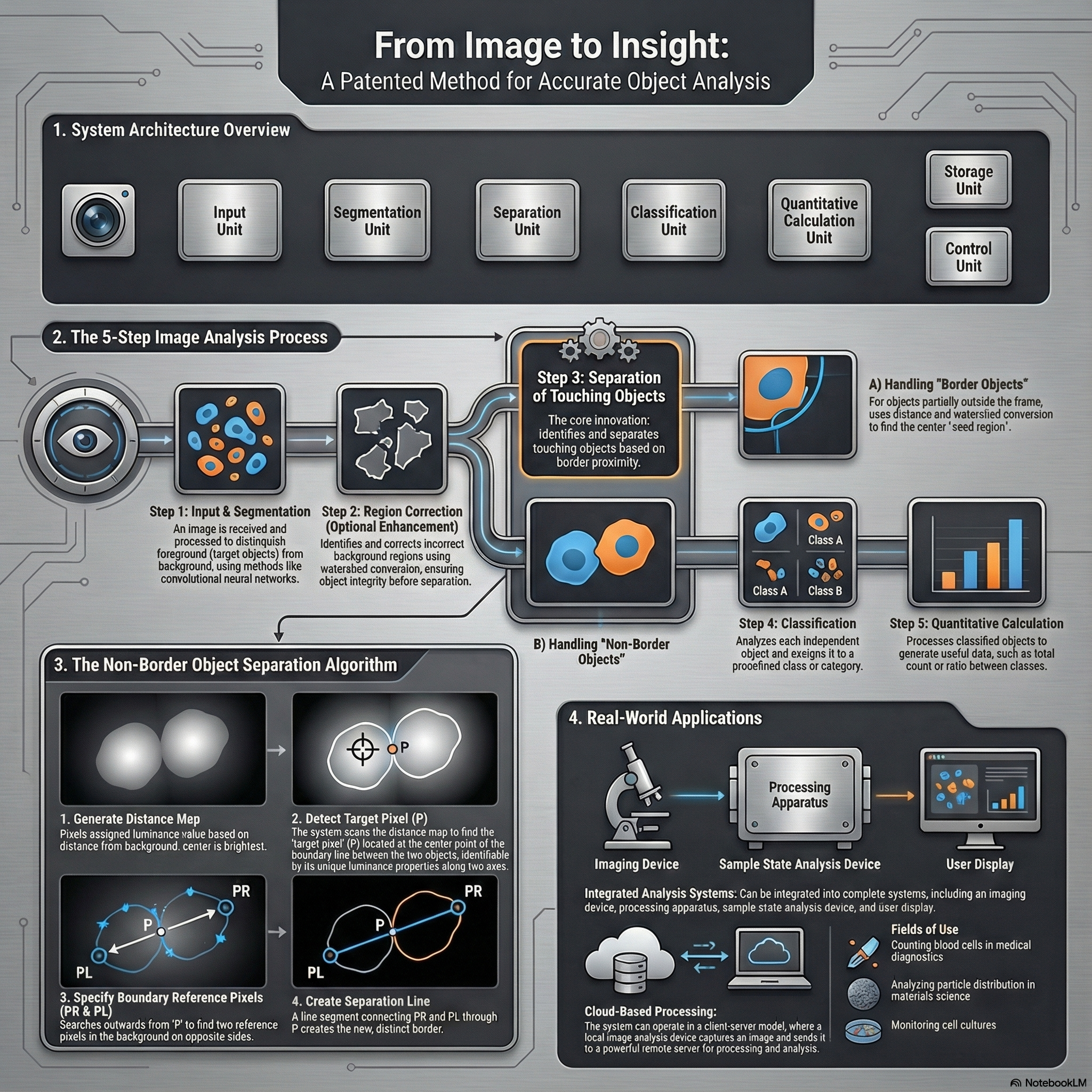
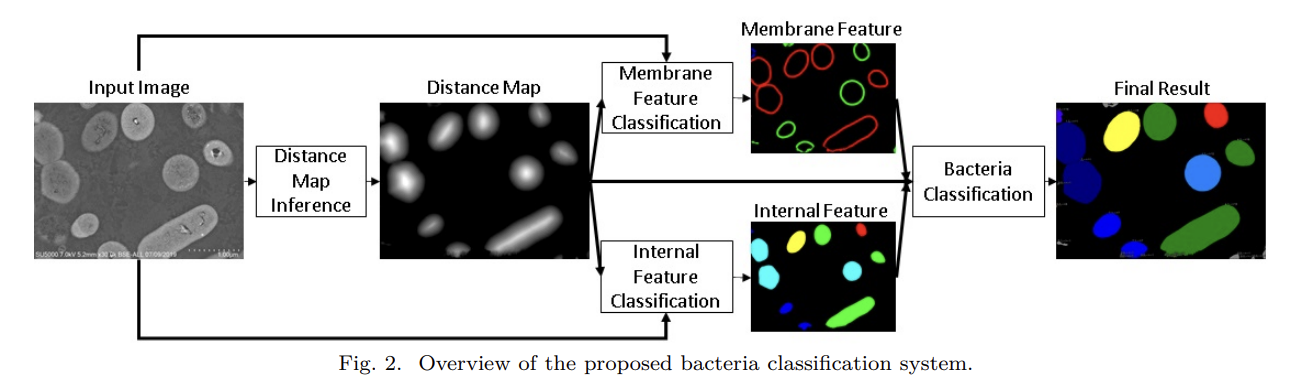
Deep Learning for Microscopy Image Analysis
From 2018 to 2021, my research at Hitachi Ltd., Tokyo focused on developing deep learning–based systems for high-precision image understanding in biomedical microscopy. I worked on combining computer vision and AI-driven automation for identifying and quantifying objects of interest in complex visual data.
Publications:
Patents:
This phase of my work established a foundation in AI-driven visual understanding, bridging industrial automation with quantitative biological imaging, and set the stage for my later research in generative and flow-based models for microscopy restoration.
Past Research (Masters Thesis, 2016–2018)
Modeling the Feature Evolution in CNNs using LSTM
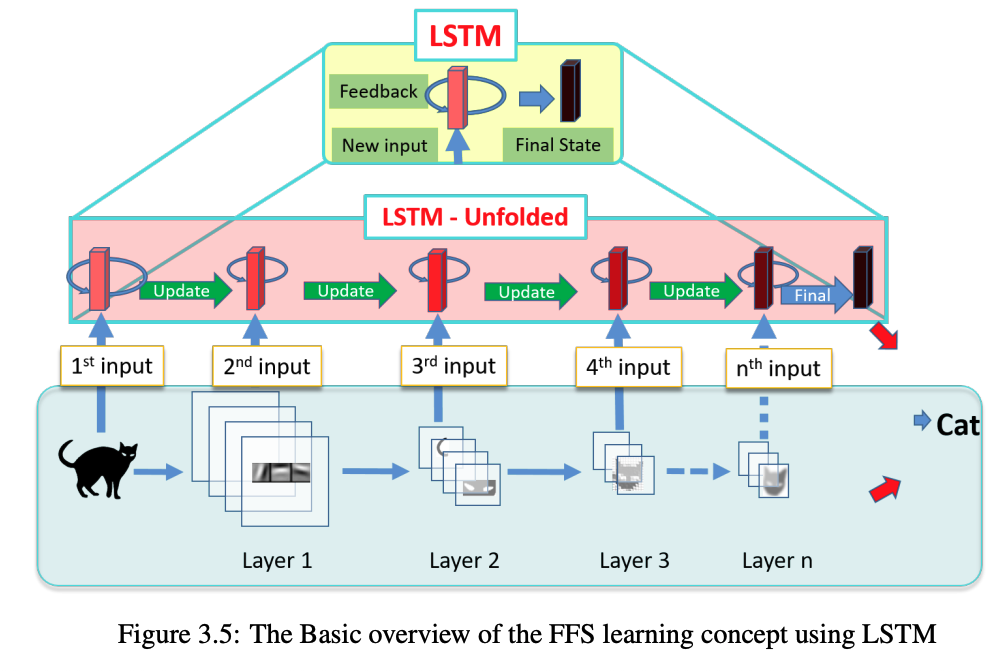
During my master’s studies at Nagoya Institute of Technology, Japan (2018), I explored the temporal dynamics of feature representations in Convolutional Neural Networks (CNNs) using Long Short-Term Memory (LSTM) networks. My research focused on understanding how features evolve across layers in CNNs and leveraging LSTMs to model these transitions for improved image classification performance. Read more about it in my thesis. | AI Generated infographic 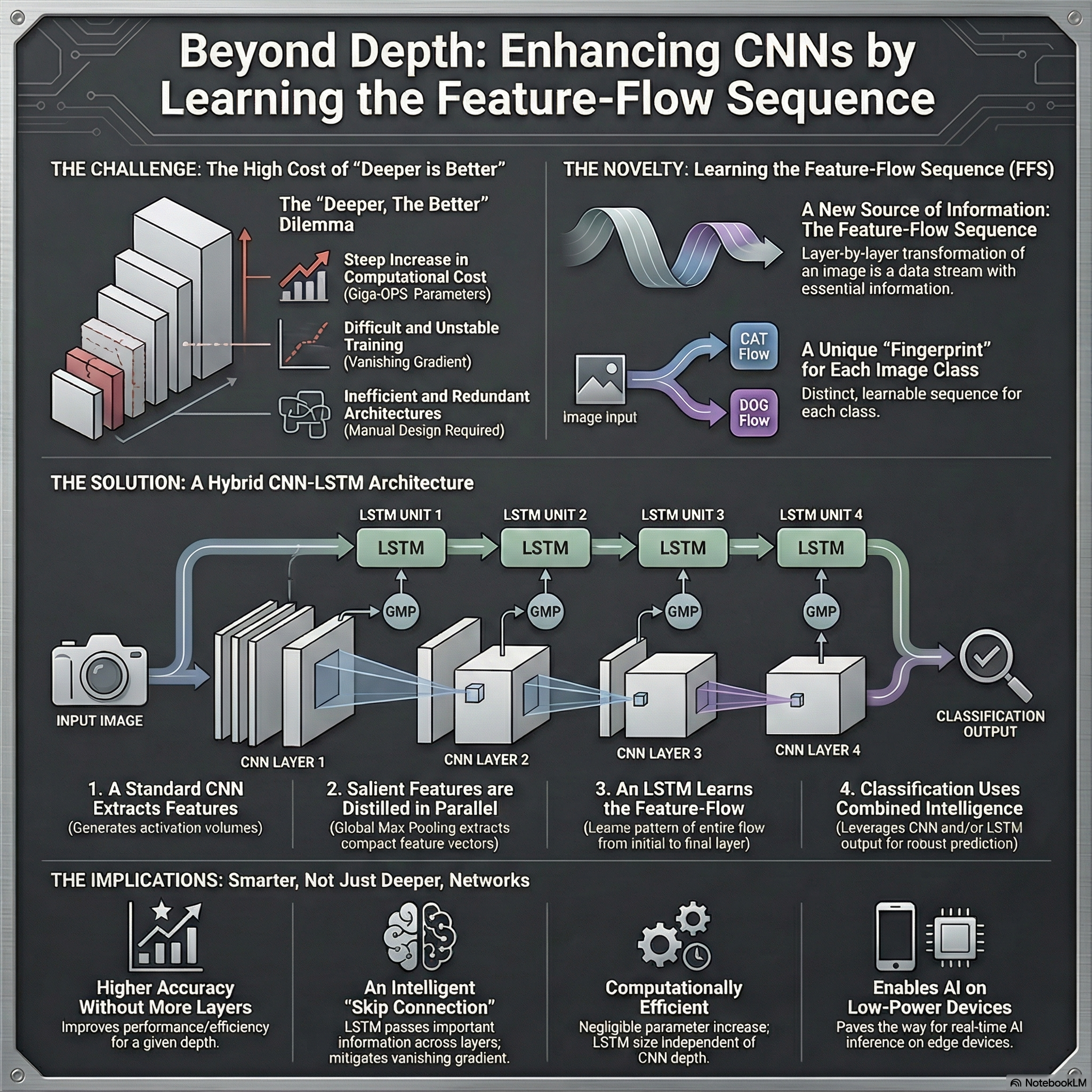
(Note that AI Generated infographics are representational only)